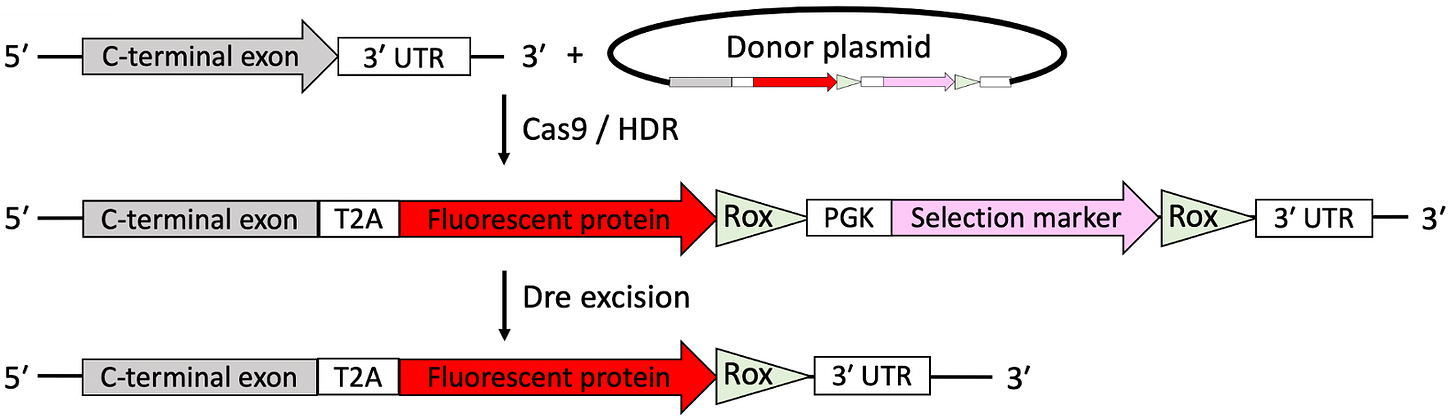
Generating gene-edited cell lines has never been easier — over the last decade, a huge proliferation of tools (CRISPR, base editing, prime editing, engineered transposases and recombinases, etc) have allowed researchers to make defined genetic changes in order to answer biological questions. In my own work, I have extensively used CRISPR to create stem cell lines with fluorescent reporter alleles for lineage-specific marker genes. Basically this involves cutting the DNA at a specific site using CRISPR and introducing a repair template plasmid that the cell integrates at the cut site. The plasmid also contains a drug selection marker, which can be removed later on by adding a recombinase.
These engineered stem cell lines will then become fluorescent when they differentiate to the correct cell type.
But sometimes things can go wrong during the editing process: off-target mutations, unwanted mutations at the on-target site, chromosomal abnormalities, microbial contamination,1 or cell line misidentification. Detecting these problems is critical for ensuring successful and reproducible research. Notably, all of these problems involve either unwanted changes to DNA sequences, or presence of unwanted DNA sequences. This means that whole genome sequencing (WGS) can, in principle, detect them.
I first looked into using WGS for cell line quality control in 2022, when I had just made a new edited cell line and wanted to verify that it didn’t have chromosomal abnormalities. I was looking at what Thermo Fisher charged ($550) for a microarray-based karyotype,2 and I thought it was absurd, because WGS provides more information for a lower price. (At the time, research-grade WGS was about $400 per sample and now it’s even lower.) So, I decided to do WGS on my cell line. The only issue was that the existing software tools for WGS were pretty complicated to use. A few papers had used WGS for cell line quality control, but they didn’t actually publish their code.3 And previous methods were often tricky to use with edited genome sequences.
Introducing SeqVerify
Over the last two years, I’ve worked together with Valerio Pepe, a computer science undergrad, to develop our own software tool, called SeqVerify, for WGS data analysis. We also collaborated with researchers at Northwestern University (Steven Lubbe and Evangelos Kiskinis), who tested SeqVerify and compared its results to their previous in-house WGS analysis. We’re excited to announce that SeqVerify has now been published in Stem Cell Reports.
Basically, what SeqVerify does is take:
raw DNA sequencing reads
a reference genome
a list of intended edits
several optional external databases (for example, microbial contaminants)
and produce a quality control report showing if the edits are present, and if there are any problems.
Knowing that installation can often be the hardest step of using a bioinformatics package, we made SeqVerify easy to install from Bioconda using a single command:
conda install -c bioconda seqverifyOverall, SeqVerify requires only minimal command line skills, making it usable for people who aren’t bioinformaticians.
We hope that everyone making edited cell lines uses SeqVerify to confirm their results. And if you’re a potential user and want to suggest changes, head over to our Github.
Here’s to better science!
Mycoplasma bacteria are especially insidious, since they usually don’t cause visible contamination or outright cell death, and are resistant to many antibiotics. A 2015 paper found that 11% of datasets from cell lines in the NCBI sequencing database were contaminated with Mycoplasma.
Basically this is looking at a few million short pieces of DNA from different chromosomes to make sure that all the chromosomes are there and no pieces are duplicated or missing.
A typical paper in this area would provide a written description of the software packages that they used and their overall analysis steps, which is NOT the same as actual working code, especially because sometimes the papers neglect to include important details about the settings/parameters they use for their packages.