This is Part 2 of the epigenetics post series. Part 1 is here.
The year was 1984, and scientists were wondering: why is sperm, and not just eggs, necessary to make a baby mouse?
In non-mammalian species (notably several reptiles), unfertilized eggs can activate as if they were fertilized, copy their genomes, and produce viable offspring. This process is known as parthenogenesis, and scientists could induce it experimentally in mouse eggs, with one research group even reporting live offspring. But as other researchers tried it, it became clear that although parthenogenetic embryos developed to the blastocyst stage, they did not successfully grow to term, and in particular showed defects in the extra-embryonic tissues such as the placenta.
Three labs, led by Azim Surani at Cambridge, Davor Solter at the Wistar Institute, and Robin Lovell-Badge at University College London, were hard at work trying to understand why.
One hypothesis was that there was some necessary component missing from the egg cytoplasm which the sperm provided. But this was quickly ruled out, because when both the male and the female pronucleus were transplanted from normally fertilized eggs into enucleated recipient eggs, the resulting embryos developed normally.1 So what was going on?
Around the same time, the Surani and Solter labs performed basically the same experiment:
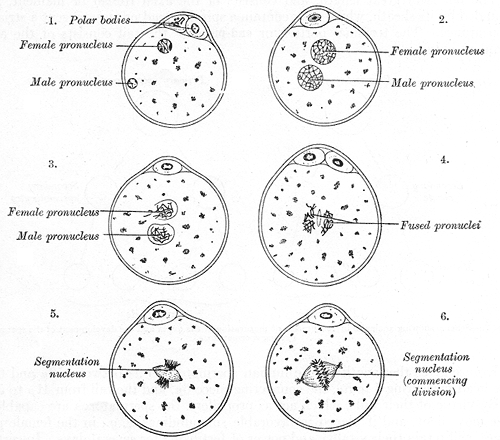
Take fertilized eggs and remove either the male or the female pronucleus using a tiny glass needle.2
Transplant a new male or female pronucleus into each egg, generating eggs with two male pronuclei, two female pronuclei, or one of each (as a control).
See how the resulting embryos developed.
Both of these research groups came to the same conclusion: the only embryos that developed to term were ones containing both a male and a female pronucleus.3 These experiments were performed using inbred mouse lines, in which an X-bearing sperm is genetically identical to an egg. So, proper development required some characteristic that was associated with the male-derived and female-derived chromosomes, but was not related to their sequences. Hmm, sounds like epigenetics.
This result led Azim Surani to remark:
Our studies suggest that specific imprinting of the genome during gametogenesis is essential for full-term development in the mouse. At some point during embryonic development and formation of primordial germ cells, previous influences on the genome are presumably lost and new ones initiated during the production of gametes.
This was a remarkable conclusion to make in 1984, but it turned out to be entirely correct.
Epigenetic imprinting
By “imprinting”, Surani meant the phenomenon where some genes are expressed from only one parent’s copy. For example, the IGF2 gene we looked at last time is expressed only from the father’s copy, not the mother’s. Other genes such as UBE3A follow the opposite pattern, being expressed only from the mother’s copy.4 In 1984, the mechanism of imprinting was unknown, but it turned out to be mediated by DNA methylation. The promoters of certain genes contain imprinting control regions that regulate gene expression depending on their methylation status5, which differs between eggs and sperm. In humans there are about 100 known imprinted protein-coding genes,6 out of a total of about 20,000.
As Surani noted, in order for imprinting to work, the parents’ imprinting marks need to be lost in the precursors of eggs or sperm, and sex-specific marks need to be written. Let’s take a closer look at how this is accomplished.
The cycle of the mammalian germline
Everything starts when a sperm fertilizes an egg, forming a zygote. The zygote undergoes a rapid series of cell divisions, forming a ball of cells called a morula. The outer cells of the morula differentiate into trophoblast cells, the origin of the placenta and other extra-embryonic tissues. These pump fluid into the center of the embryo, forming a cavity known as a blastocoel. At this point the embryo is known as a blastocyst.
The inner cell mass of the blastocyst consists of embryonic stem cells, which give rise to all the cells of the adult animal. These differentiate into distinct lineages in a process called gastrulation. Describing gastrulation in detail would fill a textbook,7 but the end result is the differentiation of the cells into three layers (ectoderm, mesoderm, and endoderm) as well as the specification of primordial germ cells (PGCs), which are the earliest precursors of eggs and sperm.
Because evolution does things weirdly, the primordial germ cells start out in the wrong place and at the wrong time. The gonads are the organs that produce gametes, but at the time of PGC specification they haven’t formed yet (they’re still just mesoderm). In a process that takes a few days in mice and a few weeks in humans, the PGCs migrate into the developing gonads. Once inside the gonads, they undergo several rounds of division, and largely erase their DNA methylation (more on this below).
The sex of the embryo is determined by the supporting cells of the gonad. If the Y-chromosomal SRY gene is present, these cells differentiate into Sertoli cells and a testis is formed. Otherwise, they become granulosa cells, forming an ovary. The germ cells follow along.8
In females, PGCs develop into oogonia, which enter meiosis, forming primary oocytes. The oocytes are surrounded by granulosa cells in structures called ovarian follicles. These follicles remain dormant for decades. During each menstrual cycle, about 10–20 follicles become activated for further growth, and in humans, only one of these follicles fully matures and releases an oocyte.9 Interestingly, the oocyte does not actually complete meiosis until after fertilization. Overall, starting from a population of a few million oogonia, only about 400 oocytes are ovulated over a woman’s reproductive lifespan. Oogenesis is very inefficient!
In males, PGCs develop into spermatogonia, which grow inside seminiferous tubules formed by Sertoli cells. Spermatogonia are unipotent stem cells of the male germline: they can either self-renew, or differentiate into spermatocytes, which undergo meiosis and further differentiate into spermatids. Spermatids are the earliest stage of cell that can fertilize an egg, but they can only do this when they are injected directly into the egg. In order to gain motility, the spermatids expel most of their cytoplasm, compact their nucleus (this has important epigenetic implications), and grow a flagellum. After completing these processes, they are now called spermatozoa. The spermatozoa travel out of the seminiferous tubules and through the epididymis, a highly coiled tube (6 meters long in humans). During this time10 the spermatozoa undergo several changes to their membrane proteins, preparing them to fertilize eggs. After passing through the epididymis, the spermatozoa are ready for release into the female reproductive tract, where they will undergo a final maturation process known as capacitation.
After all this is complete, the sperm can fertilize the egg, and the cycle can begin again.
Epigenetics of the zygote and early embryo
All humans begin as zygotes, formed by the fusion of a sperm with an egg. The sperm and egg each contribute one pronucleus, with its own pattern of epigenetic marks. But these marks change rapidly as embryonic development begins. For example, 5-methylcytosine in the paternal pronucleus is oxidatively demethylated by the enzyme TET3. Additionally, the maternal protein DPPA3 (which is only found in mammals) inhibits maintenance DNA methyltransferase activity, leading to a global drop in DNA methylation during the rapid cell divisions of the early embryo.11 Histones are also extensively replaced during this time. The overall effect is to erase most of the parental epigenetic marks, with a few important exceptions at imprinted loci.
A major developmental process taking place in the early embryo is zygotic genome activation. The oocyte contains large quantities of stored maternal mRNAs that support the first few cell divisions, but in order for the embryo to develop properly, it must switch over to mRNAs transcribed from its own genome. In humans this process is complete at around the 8-cell stage. Zygotic genome activation is associated with several epigenetic changes,12 including a broad increase in histone acetylation and changes to the patterns of histone methylation.
Epigenetics of PGCs
As Azim Surani deduced back in 1984, before sex-specific imprinting can be established, the previous imprinting must be erased. This takes place in PGCs. As PGCs proliferate in the early gonad, they replicate their DNA while shutting off the activity of the maintenance methyltransferase DNMT1. They also upregulate TET enzymes which actively remove DNA methylation (although the passive demethylation is more important). In any case, the result is a drastic drop in DNA methylation throughout the whole genome, with only about 5% of the CpG sites remaining methylated.13 For comparison, typical genome-wide methylation levels are 70–80%.
The demethylation is extensive enough that it can lead to the unwanted expression of transposable elements (TEs), which are parasitic DNA sequences that can copy themselves to new sites in the genome, causing genomic instability. To counteract this, mammals express specialized systems to repress TEs in PGCs, such as KRAB zinc-finger proteins and the piRNA silencing pathway. Additionally, some TEs remain methylated. Indeed, of the roughly 5% of CpG sites that escape demethylation in PGCs, the vast majority are associated with TEs.14
Epigenetics of mature gametes
After a near-complete erasure in PGCs, DNA methylation is re-established by the de novo methyltransferase DNMT3A as the PGCs differentiate into mature gametes. This sets up sex-specific imprinting.
In females, imprinting is established shortly after birth (at least in mice). At this time, the oocytes are arrested in the diplotene stage of meiotic prophase I, where they remain until ovulation. Meiosis involves large changes to histone marks: in particular, recombination sites have H3K4me3 added by the methyltransferase PRDM9, and after double-strand break formation these sites are also marked by phosphorylation of H2AX. Additionally, the linker histone H1 is replaced by the oocyte-specific linker histone H1FOO. The oocyte epigenome is stable during meiotic arrest, but after ovulation and resumption of meiosis, there are several complex changes to histone modifications that prepare the oocyte for fertilization.15
In males, imprinting is established when PGCs differentiate into spermatogonial stem cells, which takes place in late fetal development. Thereafter, the pattern of DNA methylation is relatively stable. However, there are substantial changes to histones as spermatogonia complete meiosis. In addition to phosphorylating H2AX at recombination sites, male germ cells also add this mark, and other repressive marks, to the histones on the un-synapsed regions of the X and Y chromosomes. This process, known as meiotic sex chromosome inactivation,16 shuts off transcription on the sex chromosomes that could otherwise disrupt meiosis. Male germ cells also express several male-specific histone variants which substitute for the normal histones.
After meiosis, histone marks change even more as the resulting spermatids differentiate into spermatozoa. Notably, mature sperm have barely any histones! In order to swim better, they replace their histones with protamines, which are small, positively charged proteins that package the DNA very tightly, causing the nucleus to compact. Protamines replace approximately 85–90% of all the histones in human sperm. However, this process is not strictly necessary for fertilizing an egg, since round spermatids are capable of producing live offspring if injected directly into eggs.
Conclusions
The cycle of mammalian reproduction involves several critical epigenetic events. In the fertilized egg (zygote), an initial round of epigenetic erasure activates the zygotic genome, but does not completely erase parent-specific imprinting of important developmental genes. Later, these imprints are thoroughly erased in primordial germ cells. During gametogenesis, new sex-specific epigenetic marks are established in preparation for the next generation.
For my own area of research (in vitro gametogenesis) it is critical to understand these processes and get them right, since improper imprinting can lead to developmental defects. If anyone ever wants to make eggs from men or sperm from women, this will be particularly important. Understanding the epigenetics of the germline and how to alter them has already led to several remarkable advances in mice, such as the creation of viable offspring from a single oocyte by artificially writing the male imprinting marks.17 But there’s still a lot we don’t know, especially for humans where it’s difficult to obtain fetal germ cells for analysis.

For transgenerational epigenetic inheritance, these processes are a formidable barrier. Disruptions to the establishment of epigenetic marks in germ cells during fetal development can certainly affect the next generation, but it’s unlikely that any such epigenetic change would be stably maintained over multiple generations.18 The only plausible way that this could happen is if the epigenetic change is in a region that escapes demethylation during PGC development, such as a transposable element.19
Next time we’ll take a closer look at the field of transgenerational epigenetic inheritance in mammals, and discuss what evidence it would take to convince me that it’s important for humans.
https://pubmed.ncbi.nlm.nih.gov/6738704/. A pronucleus is a haploid nucleus of a zygote, one of which comes from the sperm and the other from the egg.
It’s possible to tell which is which because the paternal one is slightly larger, and the maternal one is located closer to the second polar body.
The evolution of imprinting involves parental antagonism and is quite interesting, but is not the subject of this post. For more, see David Haig’s writing here.
It’s not as simple as unmethylated = higher expression, because sometimes the unmethylated allele produces a long noncoding RNA that acts as a cis-repressor of the target gene.
And indeed it does. Also, the details vary by species: for example, mouse embryos at this stage are cylindrical whereas human embryos are disk-shaped.
If there is a mismatch in sex between the germ cells and somatic cells, the germ cells will initially follow the somatic cells but then things will go wrong during meiosis.
The follicles compete with each other in a hormonal feedback mechanism, such that one becomes dominant. Sometimes this doesn’t fully work, and two oocytes are released. This gives rise to fraternal twins. During hormonal stimulation for IVF, it’s possible for all of the activated follicles to ovulate, which means 10-20 eggs can be collected (lower numbers for older women).
2-6 days in humans, 10-13 days in rodents, see: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC2639084/
https://pubmed.ncbi.nlm.nih.gov/33235224/ Interestingly if DPPA3 is expressed in non-mammalian species it also causes demethylation.
At these sites, methylation is maintained through action of DNMT3, which is probably an effector of piRNA-mediated silencing, although to my knowledge this has only been shown in slightly later stages of germline development.
I’m only claiming this for mammals. Other species do their epigenetics differently.
Indeed, agouti viable yellow mice are the only strong example of an epigenetically inherited phenotype in mammals, and it’s caused by differential methylation of a retrotransposon. And it’s still not very stable.




This is fascinating. You say mammalian, but I'm guessing the vast majority of this sort of research has been on mice and placental mammals? I'm curious how different the mechanisms are in marsupials, since they seem to handle histrones somewhat differently. I know several groups are trying to clone marsupials, and this sort of research seems like it may be relevant.
(I would not be surprised if an echidna managed to commit parthenogenesis some day. Monotremes seem to think they're still back in the Cretaceous and don't even have a SRY gene.)
Great post!